We've divided the segments into the following labels:
1. Background
2. Table
3. Main apple body
4. Shaded area
5. Stem area
6. Specular highlight
7. Cast shadow
Using the mask on the right, we can compute the histogram of each label 1-7. Using this data, we will modify the k-means clustering algorithm (setting k = 7), so that it outputs a segmentation as close to the mask on the right as possible.
Method
To do this, we will use the following equation to compute the distance D between a pixel p and a cluster A:
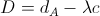.gif)
where dA is the distance between pixel p and cluster A's mean in RGB feature vector space (as per normal k-means) and c is just some parameter. We define λ as follows:
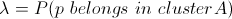
which we compute by marginalizing over our set of labels:
λc represents our confidence that we have both labeled cluster A correctly, and that pixel p belongs to that label. If this value is high, it will drive the distance D lower, increasing the likelihood of assigning pixel p to label A. If our confidence is low, λ will go to 0 and we default to using k-means. We anticipate that λ will start low but increase after several iterations.
To compute P (cluster A is label i) for each label i, we will compute the chi-squared distance between each label i and cluster A, then use the soft-max formula to convert those distances to probabilities that add up to 1. We will do the same for P (p belongs to label i), however since p is just a single pixel, its histogram will just be an impulse in each RGB channel. We will also try using the histogram of an nxn window over the pixel area as well, and compare results.
Eventually we want to compute these probabilities using more than just histogram comparisons, (using the parse graph described in the proposal) but this will be a good start for now.


.gif)
No comments:
Post a Comment